We are interested in the evolutionary cell biology, specifically in the organelles, processes and structures which are often unique among eukaryotes but represent key evolutionary innovations/adaptations. Currently, we study three unicellular organisms in which we examine novel aspects of the mitochondrial protein transport, mitochondrial dynamics and architecture. We also endeavour to understand pathogen specific processes such as the formation of the infectious cysts of intestinal parasites and the structure and biogenesis of polar tube of microsporidia - a proteinaceous nanotube through which these eukaryotes invade other eukaryotic cell.
|
The minimalist mitochondrial organelles of Giardia intestinalis.
Giardia is an anaerobic protist greatly adapted to the life in the gut. It attaches to the enterocytes via the adhesive disc and among many other peculiar features, it modified its mitochondria so they no more look like mitochondria. These organelles called mitosomes represent one of the simplest mitochondria. They lack mitochondrial genome, respiration and ATP synthesis and mediate only single metabolic role of the biosynthesis of iron-sulfur (FeS) clusters. While they reflect extreme evolutionary adaptation, Giardia mitosomes are also great experimental model for core mitochondrial processes such as (i) the synthesis and the export of FeS clusters, the organelle division and inheritence and import of cytosolic ATP to support mitosomal functions minimalist protein import machinery. |
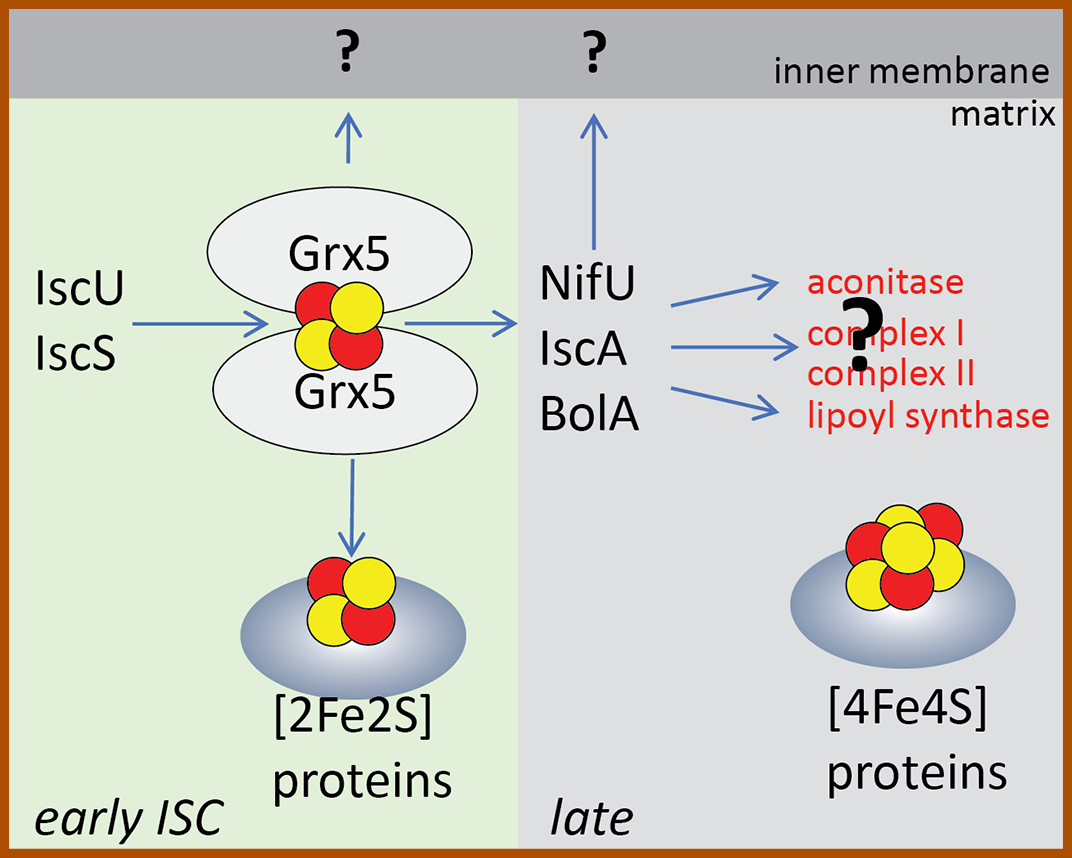
Synthesis of iron sulfur clusters in Giardia mitosome Figure i: The early ISC pathway builds [2Fe2S] clusters on glutaredoxin 5 (Grx5), the cluster is either loaded on the mitosomal apoprotein (e.g. ferredoxin) or exported as unknown molecule outside to the cytosol (question mark depicts missing Atm1 homologue). During the late ISC pathway proteins like NifU, IscA and BolA form [4Fe4S] cluster on the target apoprotein - note, that none of the known mitochondrial [4Fe4S] substrates are present in the mitosomes) (question mark depicts unknown apoprotein). |
|
Role of Mlf1 transcription factor in the quality control of mitosome and endoplasmic reticulum (ER) membranes.
Myeloid leukemia factor 1 (Mlf1) was discovered as an oncoprotein which plays role in phenotypic determination of hemopoetic cells. Surprisingly, there is Mlf1 homologue in Giardia, which can be found at various cellular membranes. We study the general role of the protein and its specific function in Giardia. |
|
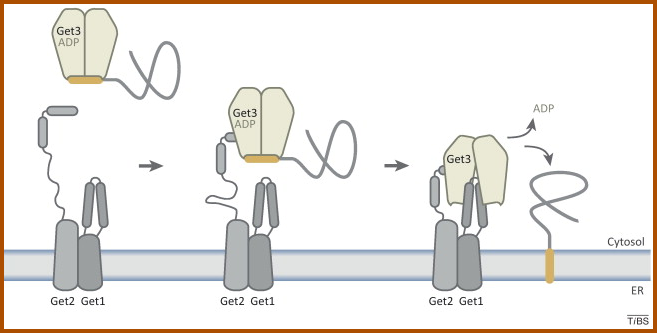
Evolution of GET pathway in eukaryotes.
Specific class of membrane proteins called tail-anchored (TA) proteins require posttranslational membrane insertion machinery. We study the evolution of this pathway using Giardia as a cellular model. |
Developing genetic tools for Giardia.
The arrival of Cas9-mediated genome editing has dramatically changed cell biology. Giardia community including our lab is trying to develop fast and reproducible approaches for genetic manipulation of tertraploid Giardia. Giardia intestinalis has two diploid nuclei and this makes reverse genetic approaches very difficult. Hence, we attempt to develop Cas9-based „single-construct approach“ capable of simultanious editing of all gene copies. |
|
Biogenesis of Giardia cysts and dynamics of encystation.
Giardia infection starts with the ingestion of the cyst stage of the parasite, from which the trophozoite stage develops in the small intestine. As little as ten cysts released from the infected animal can establish the infection. Thus the formation of the cysts is the critical virulence pathway of the parasite. We study the formation of the cyst wall and the dynamics of the overal cellular transformation. |
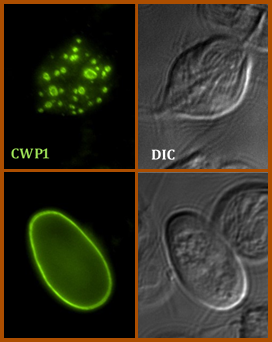
Encystation of Giardia intestinalis can be faithfully replicated in vitro.
Here, cyst wall protein 1 (CWP1, green) is secreted and deposited on the cell surface during the formation of the infectious cyst. |
|
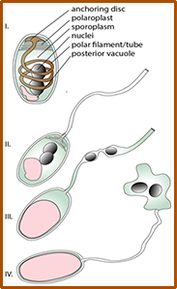
Biogenesis, composition and structure of polar tube of Microsporidia.
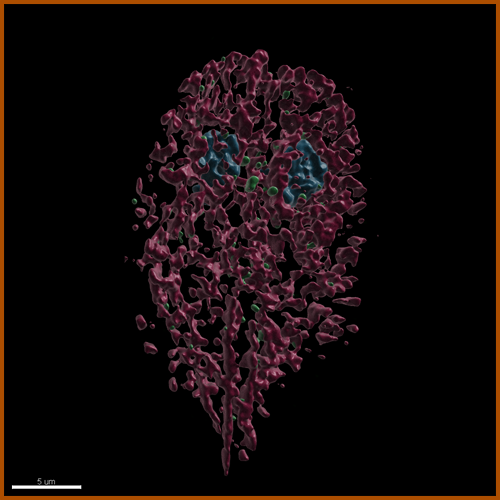
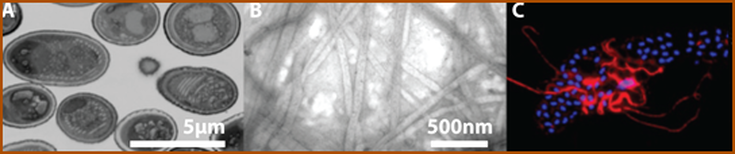
Microsporidia are radically simplified eukaryotes, with the smallest eukaryotic genomes known to date, some encoding as few as 2000 genes, yet they rely on a strikingly complex invasion apparatus centered on the polar tube. Their infectious spore ejects this proteinaceous tube through which the parasite is injected directly into the host cytoplasm. Despite being discovered in 1857 and causing serious diseases in animals including humans, Microsporidia have remained a highly neglected group of pathogens. This is mainly caused by the lack of a robust experimental model and a tractable genetic system. The biogenesis of the polar tube, its structure and evolution thus remain one of the greatest puzzles of modern biology. In this project, we study the composition, mode of action and the evolution of the polar tube. Initial characterization of N. bombycis polar tube. The spores (A) were purified on a Percoll gradient and imaged using a electron microscopy (here, as test section from FIB/SEM). The polar tubes imaged in negative stain by TEM (B). Polyclonal antibody raised against PTP2 specifically labels ejected polar tube (C) (PTP2 in red, nuclei in blue) using confocal microscopy. |
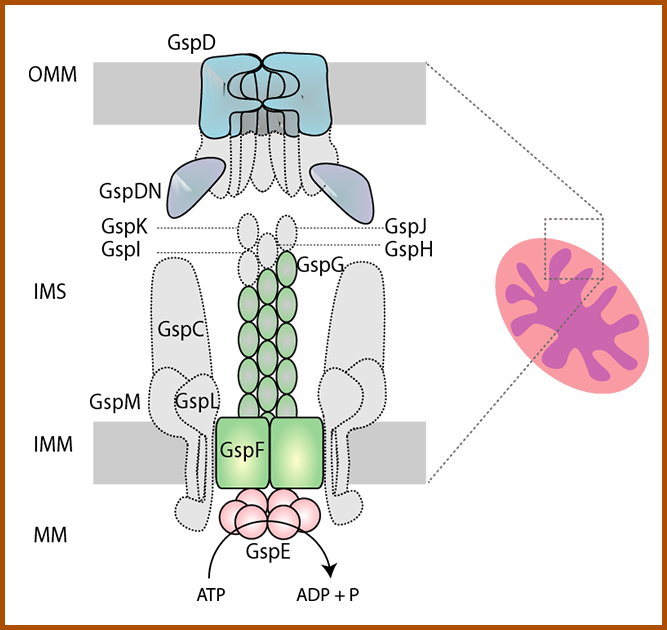
Mitochondrial protein secretion via Type II secretion system.
The acquisition of the mitochondrion was probably the trigger for the entire eukaryogenesis. The engulfment and the integration of bacterial symbiont resulted into the formation of a new cellular compartment, which retained its double membrane and became the centre of cellular energy metabolism. Recently, we have succeeded in identifying components of bacterial type 2 secretion system in mitochondria of several lineages of unicellular eukaryotes (protists). The phylogenetic distribution of the pathway suggests that the mitochondrion of the LECA was capable of secreting protein(s) to the cell cytosol. We endeavour to characterize the function of the secretion pathway and to identify proteins (and other biomolecules) secreted by protist mitochondria.
The acquisition of the mitochondrion was probably the trigger for the entire eukaryogenesis. The engulfment and the integration of bacterial symbiont resulted into the formation of a new cellular compartment, which retained its double membrane and became the centre of cellular energy metabolism. Recently, we have succeeded in identifying components of bacterial type 2 secretion system in mitochondria of several lineages of unicellular eukaryotes (protists). The phylogenetic distribution of the pathway suggests that the mitochondrion of the LECA was capable of secreting protein(s) to the cell cytosol. We endeavour to characterize the function of the secretion pathway and to identify proteins (and other biomolecules) secreted by protist mitochondria.
|
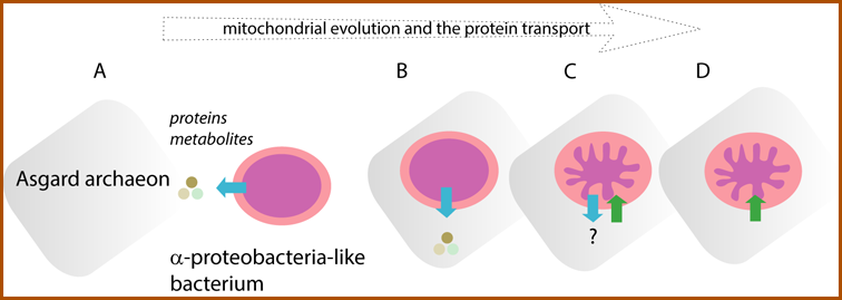
During the transformation of the bacterial symbiont into the mitochondrion, the organelle lost the capacity to secrete proteins. From the protein transport perspective, the free-living (A) and the endosymbiotic bacterium (B) was secreting proteins to the environment or to host cell cytosol, respectively. (C) Upon the transfer of the bacterial genes into host cell genome and because new functions were assigned to the organelle, proteins started to be imported from the host cell cytosol. (D) the protein export was thought to be lost from mitochondria (until now!). The export and import of proteins is depicted by blue and green arrows, respectively. |
Components of the T2SS found in several protist lineages. Our preliminary data (see below) show that the pathway is localized in the mitochondria. The proteins identified in eukaryotes (in colour) represent a functional core of T2SS and may constitute a functional mitochondrial protein secretion machine retained after the bacterial ancestor. The bacterial components missing in eukaryotes are shown in grey. MM – mitochondrial matrix, IMM – inner mitochondrial membrane, IMS, intermembrane space, OMM-outer mitochondrial membrane |